Advantages
- Improved display efficiency and post-transcriptional diversity
- Versatile and independent of functional molecules
- Established method for selecting specific RNA sequences
- Applicable to other RNA display methods
 |
Current Stage and Key Data
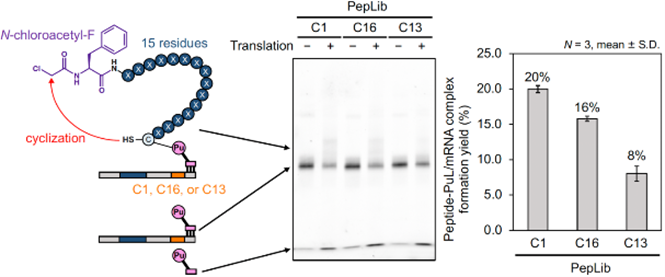
- The effectiveness of TRAP display has been confirmed using monobodies and cyclic peptides as functional molecule candidates.
- Improve display efficiency by a few percent to around 20%.
 |
Partnering Model
License this technology to companies using TRAP or other RNA display technologies.
- Potential partners: Pharmaceutical/biotech companies and CROs focusing on the discovery/development of peptide, medium molecule, and antibody drugs
Background
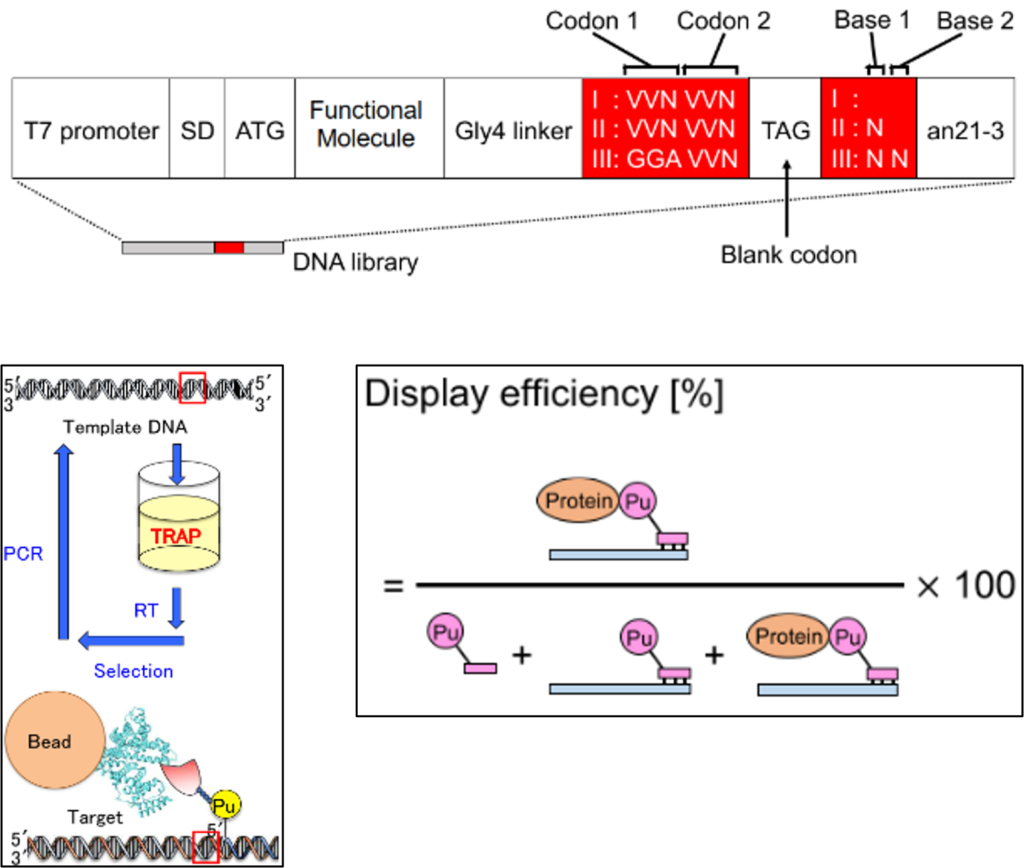
Functional molecules (peptides and proteins) with high affinity for a target are candidates for detection tools and therapeutic drugs. RNA display methods, including the TLAP method, are efficient screening methods for functional molecules by displaying phenotypes of functional molecules on mRNA (genetic type) via puromycin linker ( PuL ) based on functional molecule DNA libraries. Therefore, it is important to ensure the diversity of functional molecule-displaying mRNA based on the diversity of the DNA library.
To ensure diversity in TRAP display, it is necessary to improve the display efficiency. Three factors determine the display efficiency: the binding efficiency of mRNA and PuL , the translation efficiency of mRNA to protein, and the ligation efficiency of protein and PuL. We worked to improve this efficiency.
Patents and Publications
- Umemoto et al., Nucleic Acids Research (2023) Volume 51, Issue 14, P. 7465–7479
- Japan: Patent registered (No. 7654312), USA: Patent pending (Pub No. US2025017770A1)
Principal Investigator
Hiroshi Murakami (Graduate School of Engineering, Nagoya University, Tokai National Higher Education and Research System)
Project ID:BK-03990


