Advantages
- Proven in derivative discovery for infectious disease drugs such as polymyxin B
- Seamless integration process of “structure exploration” and “compound evaluation”
- Two patterns of “original amino acid units” and “modified peptide libraries” can be selected based on application
Background & Technology
Peptide drug development requires the examination of structural modifications to enhance activity or confer stability, but the current challenge is the enormous time and cost involved in the development process. Conventionally, the process involves first identifying the modifiable sites on the parent peptide, then preparing selectively chemically modified peptides at the identified sites, and finally evaluating them separately.
The researchers have developed an amino acid unit that allows for both the identification of modifiable sites on the parent peptide and selective chemical modification at those sites. Two patterns of this amino acid unit are available:
Process 1: After peptide scanning, the alcohol side chain of this amino acid unit undergoes serine/threonine ligation (54 synthesized compounds already created), enabling continuous evaluation of scanning and peptide chemical modification.
Process 2: Using the original amino acid unit, a wide range of commercially available aldehyde compounds can be screened and evaluated in a single step on the same plate where the modification site was scanned.
Both processes 1 and 2 allow chemical modification to be performed on a micro-scale (hundreds of nmol), achieving reductions in time and cost for synthesizing numerous peptides. This technology is believed to contribute significantly to future peptide drug discovery.
Data
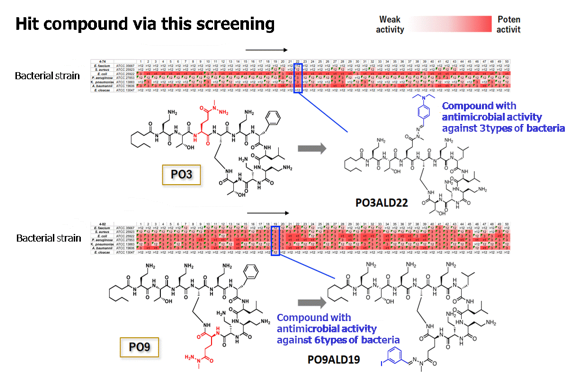
For polymyxin B, a broad substrate range was discovered through the reaction in Process 2. From 50 derivatives, the synthesis and evaluation of the derivatives were carried out with a scan of only a few milligrams using in situ screening.
The library contained many compounds exhibiting antimicrobial activity against two or more bacterial strains. Among these, P03 demonstrated antimicrobial activity against E. coli plus three other bacterial species, and P09 showed antimicrobial activity against E. coli plus six other bacterial species.
These results demonstrated that this process can be used to search for compounds with
higher antimicrobial activity than the parent peptide, as well as compounds exhibiting antimicrobial activity against a variety of bacterial species, and those showing antimicrobial activity against a large number of bacterial species.
 |
Patent
Pending Process1: international phase, Process2: domestic phase
Researcher
Hokkaido University Center for research for education on drug discovery
Professor Satoshi Ichikawa, Lecturer Akira Katsuyama
Development Phase
Current Stage: Concept validation completed for two patterns of “Original Amino Acid Unit + Library-Based Exploration Process.”
Next Phase:
- Development and evaluation of compounds identified through the search process
- Broad-range derivative search and evaluation using target peptides via this process
- Kit development and automation of this process
We are seeking partner companies interested in technology licensing or development collaboration/commercialization regarding the above. We would be pleased to begin with a detailed technical explanation and discussion.
Project ID:ON-05366


