Advantages
- Superior prediction accuracy compared to conventional target molecule prediction methods.
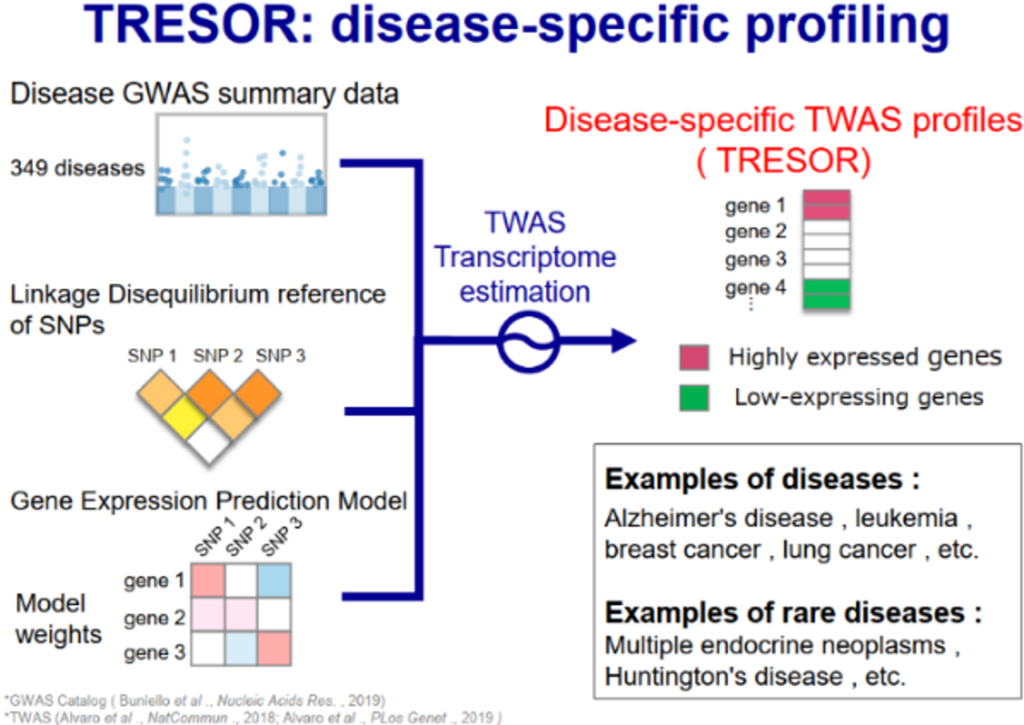
- Predictions are possible even for rare diseases with no known therapeutic target molecules.
- Therapeutic target molecule candidates have already been predicted for over 300 diseases.
 |
Current Stage and Key Data
- The validity of new target molecules predicted by this system for tauopathy, etc. has been verified using large independent cohorts.
- A database of therapeutic target molecule candidates predicted for over 300 diseases.
Partnering Model
Joint research and development of new therapeutic target molecule discovery using this system / License of know-how for therapeutic target molecule prediction algorithm / License of technology for this system
- Potential partners: biotech/pharmaceutical companies, AI-based drug discovery companies.
Background
Identifying therapeutic target molecules for diseases is a critical step in drug discovery. However, the depletion of viable therapeutic target molecules has become a major bottleneck, particularly in the field of rare and orphan diseases. The main approaches to predicting therapeutic target molecules include the use of single nucleotide polymorphisms (SNPs), genome-wide association studies (GWAS), and transcriptome data, where protein-coding genes with disease-associated SNPs or genes/proteins whose expression is altered in disease patients are selected as therapeutic target molecules. However, the number of therapeutic target candidates predicted by these methods is vast and heterogeneous, making it difficult to narrow down effective therapeutic target molecules.
Principal Investigator
Yoshihiro Yamanishi (Graduate School of Informatics, Nagoya University, Tokai National University Organization).
Reference
- Paper: Namba et al., Nat Commun . (2025) 16(1):3355.
- Patent: Patent pending (unpublished)
*Unpublished information can be disclosed under the CDA.
Project ID:BK-05201


