Summary
- A new algorithm that greatly reduces the computational cost of cluster analysis.
- The algorithm proposes a special principal component analysis. It reduces the number of features used for clustering to less than 1% of the conventional number.
- Despite the large reduction in the number of features handled, the accuracy of cluster analysis remains high.
- Our technology contributes to significant cost reduction in big data analysis within a company.
Background and Technology
Cluster analysis is becoming increasingly popular in data science, but there is a practical problem. The demand for more accurate and higher-resolution analysis has increased the number of features and samples to be measured, and the overall data volume has exploded. In other words, it is necessary to perform better analysis while reducing the cost of clustering (computer hardware, energy consumption, time, and operation and maintenance costs).
A research group led by Professor Tetsuya Sakurai of the Artificial Intelligence Science Center at the University of Tsukuba has developed a new clustering method that significantly reduces the number of features handled without degrading the accuracy of the analysis, thereby reducing costs. The core of this method is to apply a special principal component analysis to the data with a “Columbus’ egg” inversion. This process allows clustering without loss of precision, even when the number of features is reduced to about 1/10,000 of the original data. The technology developed by the University of Tsukuba is expected to solve the problem of reducing the cost of clustering processing in one fell swoop.
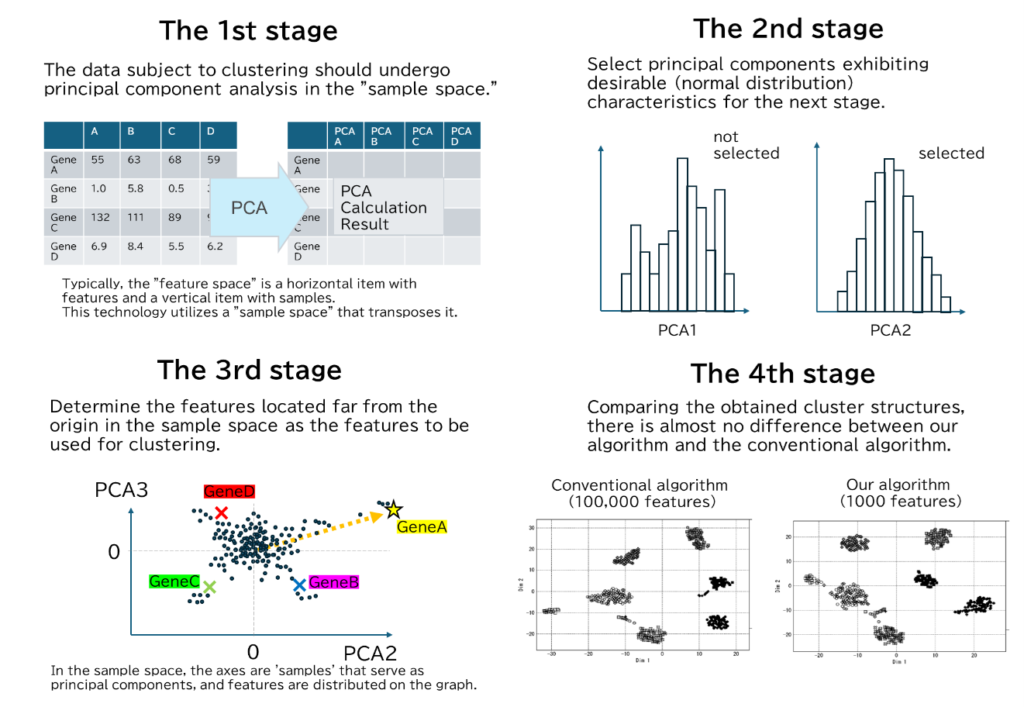
The processing of this technology is divided into four main stages. The first stage is a special principal component analysis, the second stage is the selection of principal components to be used, the third stage is the selection of feature values, and the fourth stage is clustering.
The first stage (upper left in the figure) is “special principal component analysis”. In ordinary principal component analysis (PCA), the samples are distributed on a graph with the principal component features such as the axes. This is called “PCA in feature space”. On the other hand, this technology’s principal component analysis performs PCA in “sample space,” where the rows and columns of the features and samples are transposed. In other words, the corresponding features are scattered on a graph whose axis is the “sample” which is the principal component of the obtained result.
In the second stage (top right in the figure) the principal components to be used in the final stage of processing are selected. The selection criteria are that the distribution for the principal components should be close to a normal distribution. In other words, principal components with good characteristics are selected.
In the third stage (lower left of the figure) is the features used for clustering are selected from the “PCA of the sample space” obtained in the first stage. The selection criterion is that the features should be farther from the origin when they are distributed in the sample space using the principal components selected in the second step. In other words, a certain number of features are selected in order of increasing Euclidean distance from the origin in the sample space. This number of features is arbitrary, but as mentioned above, a smaller number is sufficient than for general clustering techniques.
Finally, the fourth step is the actual clustering process. The clustering algorithm is applied as usual, but the main feature is that the number of features to be handled is determined in the third step.
As a result, the cluster structure obtained is almost the same as that of the conventional clustering algorithm. Still, the number of features handled has been reduced to about 1/100 of the conventional clustering algorithm.
Data
- The data set consists of a type of gene sequence data (a database of differences between reference gene sequences and sample sequences).
- The number of data samples is 600, and the number of features is 20 million.
- Typical unsupervised clustering algorithms (PCA, t-SNE, UMAP, etc.) handle 100,000 features.
- The algorithm of this technique, clustering, was performed with 1,000 features.
- The obtained cluster partitioning is almost the same between this technology and the conventional method (lower right of the figure). The number of features to be processed was reduced to 1/100, and the computational cost is smaller than that of the conventional method, even considering the computation required for the process of extracting features (the first to third steps described above).
 |
Publications
- Matsuda, M., Futamura, Y., Ye, X. et al. Distortion-free PCA on sample space for highly variable gene detection from single-cell RNA-seq data. Front. Comput. Sci. 17, 171310 (2023).
Patents
- WO/2022/107835
- JPA 2023-006500
Researcher
Prof. Tetsuya Sakurai (Systems and Information Sciences, University of Tsukuba)
Expectations
We believe that incorporating this technology will enhance the value of your products and services.
If your company provides data science analysis services or packaged products, we invite you to introduce this algorithm to your business and deliver a high-speed, high-precision clustering solution to your users.
Companies that utilize data science in their marketing, sales, design, manufacturing, management, etc., can expect to see significant cost reductions by leveraging this technology. We invite you to introduce this technology to your company’s data scientists. We are also available to discuss opportunities to introduce technology directly from the university.
We will assist you in the following processes with companies that have expressed interest in utilizing the technology.
- We will be happy to respond to your questions.
- Meeting with the inventor to explain details.
- Exchange of information under NDA
- Feasibility study for commercialization through joint research, etc.
- Patent licensing.
Project No.DA-04965


