Advantages
Technology Overview & Background
In response to global challenges such as population growth and energy shortages, there is an urgent need for efficient food production. Seeds, the nutrient-rich edible parts of plants, are crucial for improving the production efficiency of cereals like rice and wheat. Enlarging seeds enhances food yield, while smaller seeds offer benefits such as reduced storage space and the development of high-value varieties with improved texture.
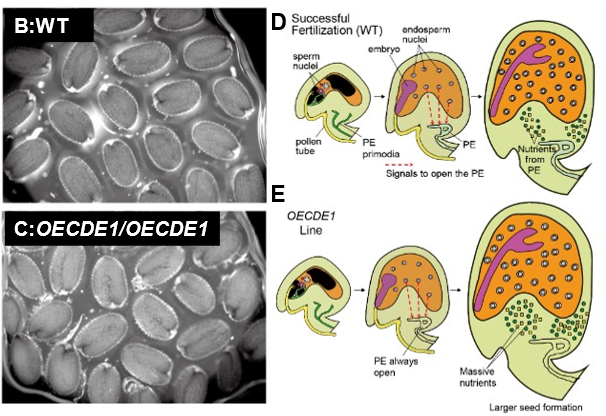
Researchers have focused on controlling seed size and discovered that overexpression of callose-degrading enzymes (CDEs) results in larger seeds. Callose, a key component of pollen and pollen tube cell walls, accumulates at the phloem end (PE) of the sieve tube and forms a structure known as the callose gate. This gate prevents the transport of hormones and nutrients to unfertilized seeds. After fertilization, the gate is gradually broken down by CDEs, allowing hormones and nutrients to reach the seeds, facilitating maturation. By enhancing CDE expression, the breakdown of callose gates and the translocation of nutrients to the seeds are increased, leading to significant seed enlargement.
The effects of CDE overexpression on seed enlargement have been confirmed in both dicotyledons and monocotyledons, with promising results for crops such as rice, wheat, soybean, and others used as food. Conversely, the loss of the CDE1 gene results in smaller seeds, indicating potential applications in producing small-seeded horticultural crops.
Data
- Rice plants overexpressing callose-degrading enzymes (Cde1OE; OEcde1/OEcde1) formed seeds that were 16% larger than the wild type. (A-C).
 |
Patent
Patent pending (Unpublished)
Principal Investigator
Dr. Ryushiro Kasahara (Nagoya University)
Expectations
The laboratory is currently attempting to create CDE gene-overexpressing rice plants by genome editing (without resorting to genetic modification) of the CDE gene promoter region in rice and is looking for companies to collaborate with. We would also like to work on controlling seed size through joint research with target plant species such as soybean, maize and sorghum. In addition to this, we can also consider other initiatives that companies are working on to control seed size in crop species.
Project.WL-04858



